2D PES Generation#
2D scans in autodE are available over distances and are optimally parallelised over the available number of cores. For example, to calculate and plot the 2D relaxed surface over the forming C-C distances in a Diels-Alder reaction between ethene and butadiene:
import autode as ade
ade.Config.n_cores = 10 # Distribute over 10 cores
# PES from the current C-C distances (~1.5 Å) to broken (3.0 Å) in 10 steps
pes = ade.pes.RelaxedPESnD(
species=ade.Molecule("cyclohexene.xyz"),
rs={(0, 5): (3.0, 10), (3, 4): (3.0, 10)},
)
pes.calculate(method=ade.methods.XTB())
pes.plot("DA_surface.png")
pes.save("DA_surface.npz")
Out:
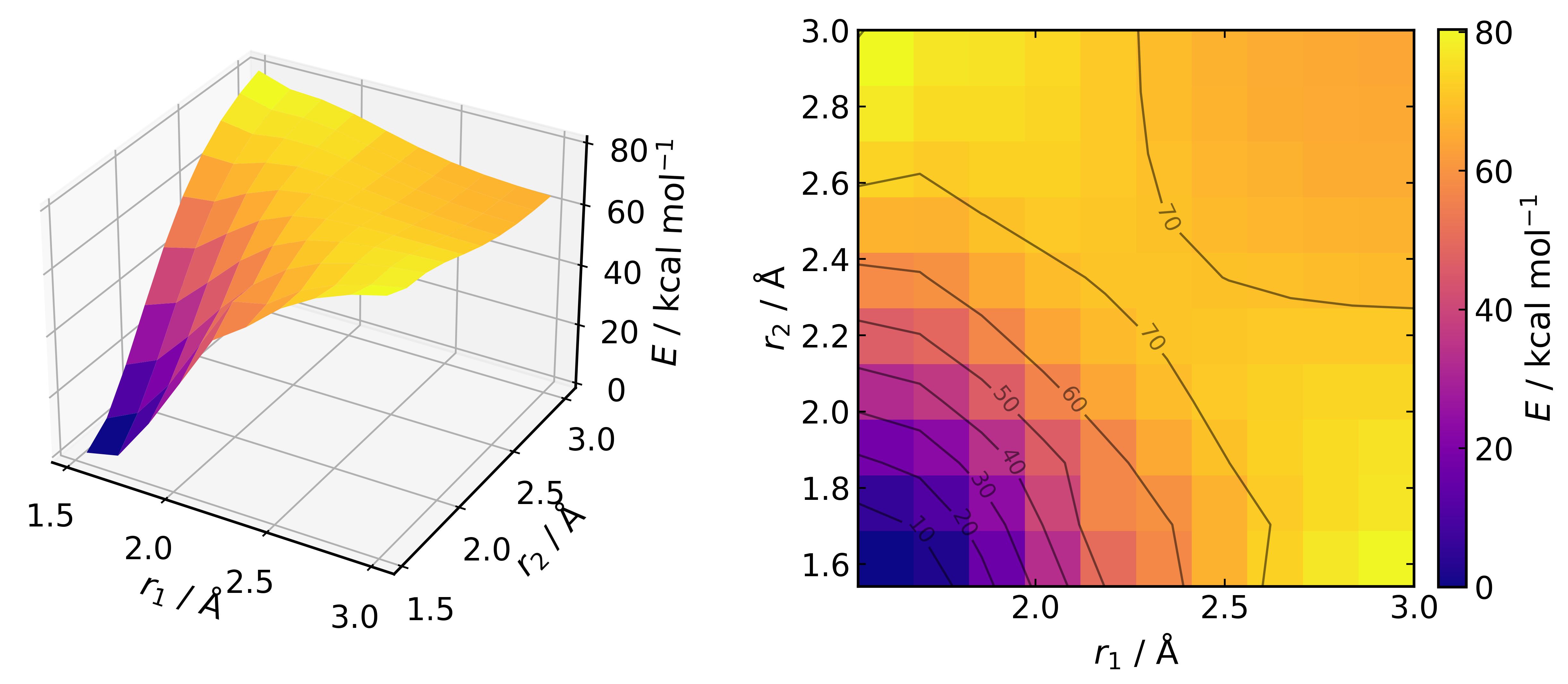
Surfaces can also be interpolated using a spline. To reload the PES and plot using a 4-fold interpolation (i.e. 10 -> 40 points in each dimension):
import autode as ade
pes = ade.pes.RelaxedPESnD.from_file("DA_surface.npz")
pes.plot("DA_surface_interpolated.png", interp_factor=4)
Out:
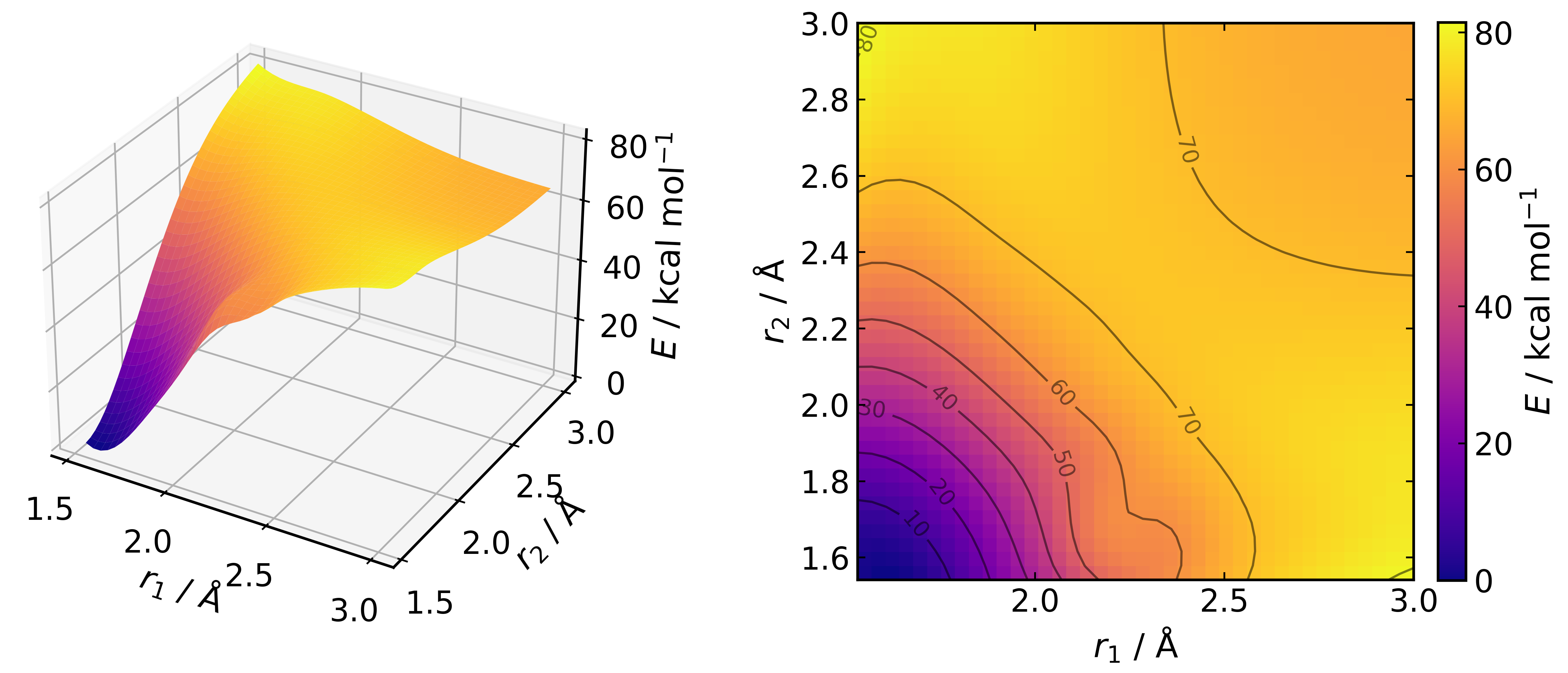
where the .xyz file (cyclohexene.xyz) used is:
16
Generated by autodE on: 2021-12-04.
C -1.25524 0.55843 -0.45127
C -0.11901 1.52914 -0.34083
C 1.12810 1.05947 -0.22343
C 1.36167 -0.42098 -0.26242
C 0.31173 -1.21999 0.53472
C -1.08411 -0.56797 0.57151
H -1.28068 0.12348 -1.46969
H -2.22404 1.06103 -0.30930
H -0.31910 2.60539 -0.34475
H 1.98105 1.73907 -0.13515
H 2.37438 -0.67767 0.08526
H 1.32299 -0.74206 -1.32088
H 0.25137 -2.23441 0.11110
H 0.67503 -1.34778 1.56617
H -1.86306 -1.33147 0.42039
H -1.26117 -0.13357 1.56876